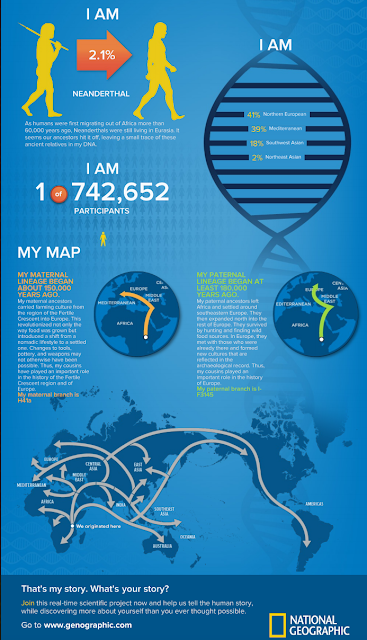
I like the idea of getting my own DNA analyzed and studying the results. In an earlier post, I described some results of an analysis through a National Geographic project, and the finding that 2.5% of my DNA was from Neanderthals. The ability to sequence ancient DNA is a relatively new capability and in the few years that it has been done, it has yielded a number of significant findings. The applicability to the field of psychiatry is limited at this time to 2 references (1,2). An additional search on ancient DNA and psychiatry yields 4 additional references (4-6) looking at the early origins of mutation and how the associated disrupted regulatory mechanisms could lead to psychopathology. One of the susceptibility markers for schizophrenia dates back to the last glacial maximum or 24,500 years BCE.
After trying out the National Geographic site, I decided to see if the 23andMe site had anything more to offer. I pulled up their web site, paid the fee and they sent me a sampling kit. Their sampling technology if different in that they use a tube of saliva as the sample rather than a scraping from the buccal mucosa. They send you a number of e-mail updates and finally a notification that your DNA has been processed and the necessary reports have been generated. There are 67 reports in all that focus on ancestry, carrier status, wellness, and traits. Some reports are more useful than others. For example it is interesting that a 4th digit on the hand longer than the second digit or a second toe longer than the great toe are inherited characteristics with certain probabilities. Unless there are some additional health implications involved I don't really care about my longest toe or finger. The data I am looking for is precisely the data that the FDA told 23andMe that it should not market in the first place. That initial FDA warning looks at the testing that the company was offering. In this document the abbreviation PGS is used for "Personal Genome Service":
I am sure there are plenty of posts around the Internet on the regulatory aspects of DNA testing and what the FDA is doing to protect the American public. 23andMe does have consistent qualifying statements saying that none of the data is for medical purposes, only for research and education. My interest is purely personal research and education. What more can I learn about DNA, genetics, molecular biology and human diseases. As noted in my original, what more can I learn about my ancestry and the fascinating subject of Paleogenetics and the associated big questions like why is Homo sapiens - the genus and species of all current human beings the only surviving Homo genus. Why are all of the others extinct? I also think that it is quite instructive to remind ourselves that as members of that species we all started out in East Africa and migrated all over the world.
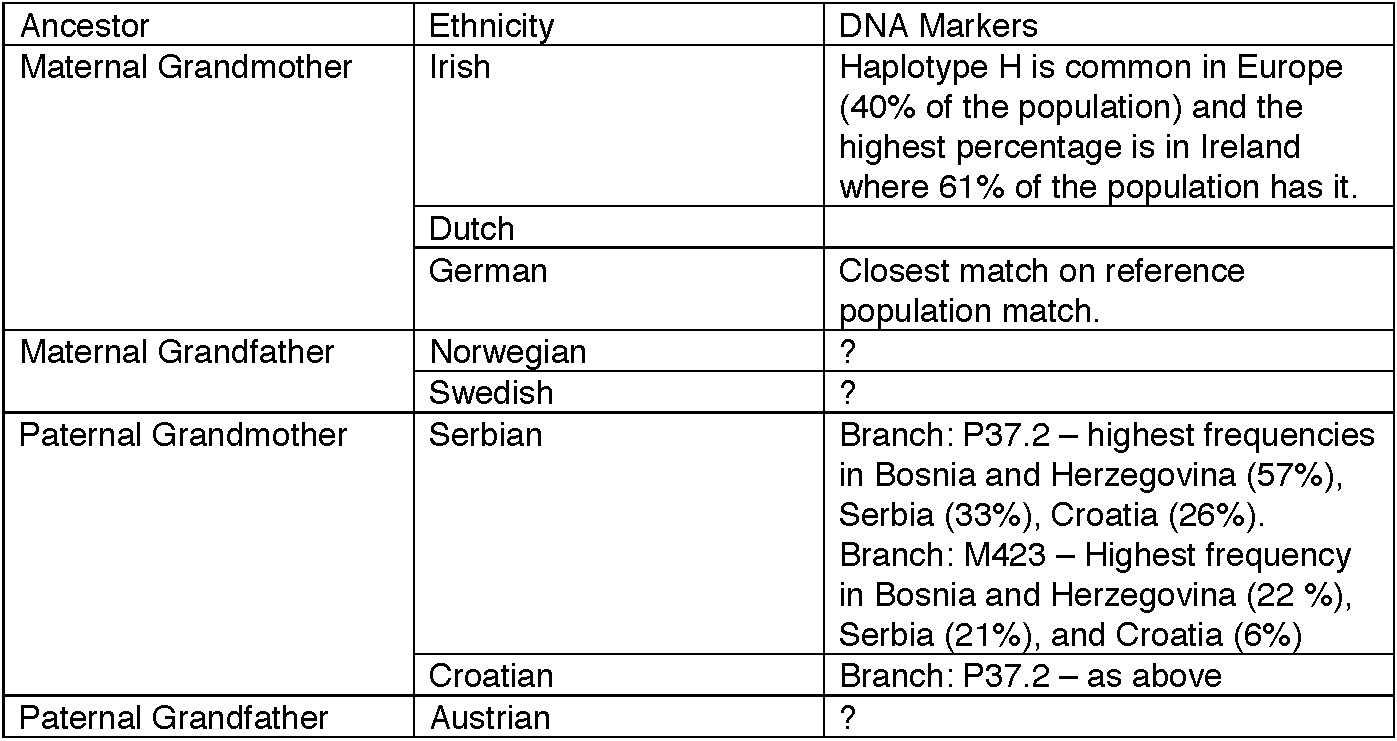
Looking at the test results from the 23and Me analysis, there are four major categories and some are more useful than others. Those categories include ancestry, traits, carrier status, and wellness. Since ancestry was the focus of the National Geographic experiment I took a look at that report first. In terms of methodology the 23and Me technology looks at overlapping regions of DNA and homology with comparison regions of known ethnic groups. I prepared the following table to look at the predictive value of the 23andMe approach compared with the National Geographic technique looking at the purported ancestries of my grandparents. (click on any graphic to enlarge)
All together there are 3 ancestry reports and the most interesting report for me was the Neanderthal analysis. The test looks at 1,436 traits across the genome and generates a report based on a map of all 23 chromosomes and a table that takes a more detailed look several markers.
The traits report was significantly less interesting to me. It answered the question about whether or not a genetic markers for a trait existed. For example, the length of the second toe on the foot and the length of the fourth finger on the hand. The testing predicts that I have a 96% chance of lighter skin and very little chance of freckling. That is true.
Carrier status was similarly not very interesting. There was a major focus on congenital illness rather than risk of chronic illness. Some of the carrier states mentioned include: ARSACS, Agenesis of the Corpus Callosum With Peripheral neuropathy(ACCPN), Autosomal Recessive Polycystic Kidney Disease (ARPKD) and 33 others. My carrier status for these relatively rare conditions was negative. That was really no surprise considering my age, family history, and the fact that most of these are illnesses of infancy or childhood.
The wellness section contained 6 reports and a few were moderately interesting. Caffeine consumption is regulated by variants near the CYP1A2 and AHR genes and I have those variants. The prevalence of these variants in various populations are also estimated and it seem that these variants are high. I do tend to consume significant amounts of caffeine and it is hardly noticeable. It seems like I am drinking decaffeinated beverages. I have the rs73598374 variant in the ADA gene that is associated with deep sleep. I do sleep for short periods of time, but my activity monitor suggests that my sleep may be deeper than people who sleep more hours. I also have the rs3923809 variant in the BTBD9 gene that predicts more movement in sleep. The R577X variant in the ACTN3 gene is present and that is associated with a greater portion of fast twitch muscle fibers or what the site refers to as sprinter/power muscle type. The final two wellness traits were lactose intolerance and the alcohol flushing reaction. I knew that I had neither trait prior to the genetic testing.
Apart from the Neanderthal testing, the most interesting aspect of the this service is that ability to search your genome looking for points of interest. I think that this will eventually be the most interesting aspect of these services as long as the users keep in mind that having a genotype, especially of a complex polygenic illness is a probability statement rather than a guarantee. I am testing out two ways to do these searches. The first strategy is based on protein analysis and I used a recent paper on bipolar disorder (I have a strong family history) to see if I could find any of these markers. The original paper suggests that there are higher plasma concentrations of 6 proteins in bipolar disorder including GDF-15, HPX, HPN, MMP-7, RBP-4, and TTR. A direct search yields a significant number of hits for HPX (5), HPN (14), and TTR (89) genes with specific information on markers, genomic position, possible variants and genotype. In this case the original paper was a protein analysis and as far as I can tell there is no genetic analysis of the subgroup with higher levels of the identified proteins. I have sent an e-mail to the lead author to see if I missed any papers on that issue. An example of the data available searching on these proteins for the HPX protein is shown below:
The second option would be to search for known genotypes. It is no secret from previous posts that I have asthma that was quiescent for most of my life that was reactivated about 3 years ago by a upper respiratory infection. Asthma is an interesting disorder because the genetics are very complex just like psychiatric disorders. For the critics who suggest that there are no tests of any sort for psychiatric disorders, these two sentences are from the latest chapter on the genetics of asthma from UpToDate (9) are instructive:
"Exploration of the genetics of asthma has also been hampered by the fact that there is no "gold standard" diagnostic test for asthma, and the clinical diagnosis is inconsistently applied. To circumvent these issues, investigators have studied the distribution of asthma-related traits, including bronchial hyperresponsiveness and measures of atopy (eg, total serum IgE levels, skin test reactivity) in addition to the presence of an asthma diagnosis."
This same author reviews the genetic research on asthma to date and points out that prior to the retirement of the Genetic Association Database in 2014 there were over 500 genetic association studies on asthma that identified hundreds of candidate genes for asthma. From those candidate gene studies, she gives the most replicated genes as filaggrin (FLG) - an epithelial barrier gene also important in atopic dermatitis, ORMDL3 - a transmembrane protein, Beta-2 adrenergic receptor gene, and Interleukin-4 receptor gene. Genome-wide association studies (GWAS) have supplanted candidate gene studies and over 50 GWAS have been done in asthma. These studies have identified other candidate genes and generally shown that GWAS done in populations with European ancestry seem to have little applicability in more ethnically diverse populations. ORMDL3 was identified in both types of studies so I searched for that in my own DNA and came up with the following:
In order to look at specific markers and asthma risk, I searched on one of the genotyped markers (rs8076131) in PubMed and came up with 7 papers on asthma susceptibility. Searching more broadly on ORMDL3 showed 132 references that were less specific.
This ends my preliminary review on the availability of personal genomics for education and research purposes by individuals. I hope to come up with more effective strategies to look at several additional disease phenotypes that I either personally possess or that were present in my first degree relatives. For me, the paleogenetics and personal genome browsing were the most interesting aspects of this data. For educational purposes, it highlights the difficulties of correlating genetics with disease phenotypes due in part to the fact that multiple genes and polygenes can produce the same phenotype and that makes the activity of specific genes difficult to determine in a DNA sample.
George Dawson, MD, DLFAPA
References:
1: Srinivasan S, Bettella F, Mattingsdal M, Wang Y, Witoelar A, Schork AJ, Thompson WK, Zuber V; Schizophrenia Working Group of the Psychiatric Genomics Consortium, The International Headache Genetics Consortium, Winsvold BS, Zwart JA, Collier DA, Desikan RS, Melle I, Werge T, Dale AM, Djurovic S, Andreassen OA. Genetic Markers of Human Evolution Are Enriched in Schizophrenia. Biol Psychiatry. 2015 Oct 21. pii: S0006-3223(15)00855-0. doi: 10.1016/j.biopsych.2015.10.009. [Epub ahead of print] PubMed PMID: 26681495.
3: Sipahi L, Uddin M, Hou ZC, Aiello AE, Koenen KC, Galea S, Wildman DE. Ancient
evolutionary origins of epigenetic regulation associated with posttraumatic
stress disorder. Front Hum Neurosci. 2014 May 13;8:284. doi:
10.3389/fnhum.2014.00284. eCollection 2014. PubMed PMID: 24860472; PubMed Central
PMCID: PMC4026723.
As noted in the above table, there is more coverage of ethnicities, using the 23andMe approach with the best example being that it picks up Norwegian, Swedish, and Dutch markers that were not present in the NatGeo analysis. There are a few problems that might not be obvious at first. The test subject does complete information about ethnicities and populations of origin that may be incorporated into the algorithm that assigns probabilities of certain ancestry. These questions reminded me of the clinical data required for quantitative electroencephalogram machines in the 1990s. The algorithms were supposed to predict psychiatric diagnoses, but the clinical data that was required with every test, frequently interfered with what the machine was going to select. I take a very dim view of what appear to be scientific decisions being made on the basis of added speculative data. The ancestry interpretations also depend the level of confidence assigned to the analysis. As an example, take a look at the following genealogy assessments - the top a conservative estimate and the bottom much more speculation. Significant changes in the analysis occur just based on how speculative the analysis is. All things considered, I am quite interested in the range of the analysis and all correlations at this point rather than precision, but is some cases precision is apparently available.
All together there are 3 ancestry reports and the most interesting report for me was the Neanderthal analysis. The test looks at 1,436 traits across the genome and generates a report based on a map of all 23 chromosomes and a table that takes a more detailed look several markers.
The traits report was significantly less interesting to me. It answered the question about whether or not a genetic markers for a trait existed. For example, the length of the second toe on the foot and the length of the fourth finger on the hand. The testing predicts that I have a 96% chance of lighter skin and very little chance of freckling. That is true.
Carrier status was similarly not very interesting. There was a major focus on congenital illness rather than risk of chronic illness. Some of the carrier states mentioned include: ARSACS, Agenesis of the Corpus Callosum With Peripheral neuropathy(ACCPN), Autosomal Recessive Polycystic Kidney Disease (ARPKD) and 33 others. My carrier status for these relatively rare conditions was negative. That was really no surprise considering my age, family history, and the fact that most of these are illnesses of infancy or childhood.
The wellness section contained 6 reports and a few were moderately interesting. Caffeine consumption is regulated by variants near the CYP1A2 and AHR genes and I have those variants. The prevalence of these variants in various populations are also estimated and it seem that these variants are high. I do tend to consume significant amounts of caffeine and it is hardly noticeable. It seems like I am drinking decaffeinated beverages. I have the rs73598374 variant in the ADA gene that is associated with deep sleep. I do sleep for short periods of time, but my activity monitor suggests that my sleep may be deeper than people who sleep more hours. I also have the rs3923809 variant in the BTBD9 gene that predicts more movement in sleep. The R577X variant in the ACTN3 gene is present and that is associated with a greater portion of fast twitch muscle fibers or what the site refers to as sprinter/power muscle type. The final two wellness traits were lactose intolerance and the alcohol flushing reaction. I knew that I had neither trait prior to the genetic testing.
Apart from the Neanderthal testing, the most interesting aspect of the this service is that ability to search your genome looking for points of interest. I think that this will eventually be the most interesting aspect of these services as long as the users keep in mind that having a genotype, especially of a complex polygenic illness is a probability statement rather than a guarantee. I am testing out two ways to do these searches. The first strategy is based on protein analysis and I used a recent paper on bipolar disorder (I have a strong family history) to see if I could find any of these markers. The original paper suggests that there are higher plasma concentrations of 6 proteins in bipolar disorder including GDF-15, HPX, HPN, MMP-7, RBP-4, and TTR. A direct search yields a significant number of hits for HPX (5), HPN (14), and TTR (89) genes with specific information on markers, genomic position, possible variants and genotype. In this case the original paper was a protein analysis and as far as I can tell there is no genetic analysis of the subgroup with higher levels of the identified proteins. I have sent an e-mail to the lead author to see if I missed any papers on that issue. An example of the data available searching on these proteins for the HPX protein is shown below:
The second option would be to search for known genotypes. It is no secret from previous posts that I have asthma that was quiescent for most of my life that was reactivated about 3 years ago by a upper respiratory infection. Asthma is an interesting disorder because the genetics are very complex just like psychiatric disorders. For the critics who suggest that there are no tests of any sort for psychiatric disorders, these two sentences are from the latest chapter on the genetics of asthma from UpToDate (9) are instructive:
"Exploration of the genetics of asthma has also been hampered by the fact that there is no "gold standard" diagnostic test for asthma, and the clinical diagnosis is inconsistently applied. To circumvent these issues, investigators have studied the distribution of asthma-related traits, including bronchial hyperresponsiveness and measures of atopy (eg, total serum IgE levels, skin test reactivity) in addition to the presence of an asthma diagnosis."
This same author reviews the genetic research on asthma to date and points out that prior to the retirement of the Genetic Association Database in 2014 there were over 500 genetic association studies on asthma that identified hundreds of candidate genes for asthma. From those candidate gene studies, she gives the most replicated genes as filaggrin (FLG) - an epithelial barrier gene also important in atopic dermatitis, ORMDL3 - a transmembrane protein, Beta-2 adrenergic receptor gene, and Interleukin-4 receptor gene. Genome-wide association studies (GWAS) have supplanted candidate gene studies and over 50 GWAS have been done in asthma. These studies have identified other candidate genes and generally shown that GWAS done in populations with European ancestry seem to have little applicability in more ethnically diverse populations. ORMDL3 was identified in both types of studies so I searched for that in my own DNA and came up with the following:
In order to look at specific markers and asthma risk, I searched on one of the genotyped markers (rs8076131) in PubMed and came up with 7 papers on asthma susceptibility. Searching more broadly on ORMDL3 showed 132 references that were less specific.
This ends my preliminary review on the availability of personal genomics for education and research purposes by individuals. I hope to come up with more effective strategies to look at several additional disease phenotypes that I either personally possess or that were present in my first degree relatives. For me, the paleogenetics and personal genome browsing were the most interesting aspects of this data. For educational purposes, it highlights the difficulties of correlating genetics with disease phenotypes due in part to the fact that multiple genes and polygenes can produce the same phenotype and that makes the activity of specific genes difficult to determine in a DNA sample.
George Dawson, MD, DLFAPA
References:
1: Srinivasan S, Bettella F, Mattingsdal M, Wang Y, Witoelar A, Schork AJ, Thompson WK, Zuber V; Schizophrenia Working Group of the Psychiatric Genomics Consortium, The International Headache Genetics Consortium, Winsvold BS, Zwart JA, Collier DA, Desikan RS, Melle I, Werge T, Dale AM, Djurovic S, Andreassen OA. Genetic Markers of Human Evolution Are Enriched in Schizophrenia. Biol Psychiatry. 2015 Oct 21. pii: S0006-3223(15)00855-0. doi: 10.1016/j.biopsych.2015.10.009. [Epub ahead of print] PubMed PMID: 26681495.
2: Mariotti M, Smith TF, Sudmant PH, Goldberger G. Pseudogenization of testis-specific Lfg5 predates human/Neanderthal divergence. J Hum Genet. 2014 May;59(5):288-91. doi: 10.1038/jhg.2014.6. Epub 2014 Mar 6. PubMed PMID: 24599118.
4: Zhang W, Tang J, Zhang AM, Peng MS, Xie HB, Tan L, Xu L, Zhang YP, Chen X, Yao
YG. A matrilineal genetic legacy from the last glacial maximum confers
susceptibility to schizophrenia in Han Chinese. J Genet Genomics. 2014 Jul
20;41(7):397-407. doi: 10.1016/j.jgg.2014.05.004. Epub 2014 Jun 2. PubMed PMID:
25064678.
5: Cotney J, Muhle RA, Sanders SJ, Liu L, Willsey AJ, Niu W, Liu W, Klei L, Lei
J, Yin J, Reilly SK, Tebbenkamp AT, Bichsel C, Pletikos M, Sestan N, Roeder K,
State MW, Devlin B, Noonan JP. The autism-associated chromatin modifier CHD8
regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015
Mar 10;6:6404. doi: 10.1038/ncomms7404. PubMed PMID: 25752243; PubMed Central
PMCID: PMC4355952.
6: Toyota T, Yoshitsugu K, Ebihara M, Yamada K, Ohba H, Fukasawa M, Minabe Y,
Nakamura K, Sekine Y, Takei N, Suzuki K, Itokawa M, Meerabux JM, Iwayama-Shigeno
Y, Tomaru Y, Shimizu H, Hattori E, Mori N, Yoshikawa T. Association between
schizophrenia with ocular misalignment and polyalanine length variation in PMX2B.
Hum Mol Genet. 2004 Mar 1;13(5):551-61. Epub 2004 Jan 6. PubMed PMID: 14709596.
7: FDA Warning Letter to 23andMe
8: Frye MA, Nassan M, Jenkins GD, Kung S, Veldic M, Palmer BA, Feeder SE, Tye SJ, Choi DS, Biernacka JM. Feasibility of investigating differential proteomic expression in depression: implications for biomarker development in mood disorders. Transl Psychiatry. 2015 Dec 8;5:e689. doi: 10.1038/tp.2015.185. PubMed PMID: 26645624.
9: Barnes KC. Genetics of Asthma. In: UpToDate, Barnes PJ, Raby BA, Hollingsworth H (Ed), UpToDate, Waltham, MA. (Accessed on April 8, 2016)
Attributions:
7: FDA Warning Letter to 23andMe
8: Frye MA, Nassan M, Jenkins GD, Kung S, Veldic M, Palmer BA, Feeder SE, Tye SJ, Choi DS, Biernacka JM. Feasibility of investigating differential proteomic expression in depression: implications for biomarker development in mood disorders. Transl Psychiatry. 2015 Dec 8;5:e689. doi: 10.1038/tp.2015.185. PubMed PMID: 26645624.
9: Barnes KC. Genetics of Asthma. In: UpToDate, Barnes PJ, Raby BA, Hollingsworth H (Ed), UpToDate, Waltham, MA. (Accessed on April 8, 2016)
Attributions:
All of the above graphics and tables with the sole exception the the ancestry table were generated with on site software at 23andMe based on my personal DNA sample.






.png)